Li, C.-F., Lin, J., Kulhanek, D.K., and the Expedition 349 Scientists
Proceedings of the International Ocean Discovery Program Volume 349
publications.iodp.org
https://doi.org/10.14379/iodp.proc.349.202.2018
Data report: exploring the presence of Bdellovibrio and like organisms in deep-sea sediment by culture-independent and culture-dependent methods1
Henry N. Williams,2 Nan Li,3 and the Expedition 349 Scientists4
Keywords: International Ocean Discovery Program, IODP, JOIDES Resolution, Expedition 349, South China Sea Tectonics, Site U1433, Site U1435, microbiology, Bdellovibrio-like organisms, BALOs, predatory bacteria
MS 349-202: Received 15 December 2016 · Accepted 14 May 2018 · Published 25 July 2018
Abstract
Given the diversity of bacteria in the deep ocean, it is logical to assume that among them are bacterial predators that play a role in cycling of nutrients and structuring of prey communities, just as they do in other ecosystems. Although bacterial predators are commonly found in the open ocean, there have not been targeted studies on them in the subseafloor. The goal of this study was to investigate the presence, diversity, and distribution of the predatory bacteria Bdellovibrio and like organisms (BALOs) in subseafloor sediments from the South China Sea. Our specific objectives were to detect BALOs in deep-sea sediment and basalt collected during International Ocean Discovery Program Expedition 349 using molecular and culture methods to test the susceptibility of both deep-sea and marine bacteria, including human pathogens, to the recovered isolates, and to determine the similarity of newly discovered deep-sea predators to known isolates through 16S rRNA sequences cataloged in GenBank. Although previously detected by others using culture-independent methods in other settings, BALOs were not detected by either culture-dependent or culture-independent methods in this preliminary investigation.
Introduction
Recent explorations of the deep ocean floor have revealed diverse microbial communities (Marchesi et al., 2001; Jørgensen and Boetius, 2007). The functions of these microbes are just beginning to be investigated but are likely as diverse as the organisms themselves. Given the diversity of bacteria in ocean and subsurface, it is logical to assume that among them are bacterial predators that play a role in bacterial mortality, cycling of nutrients, and structuring of prey communities, just as they do in other ecosystems (Hagström et al., 1988). Predation is an important universal process among many communities of organisms. Many bacteria in the oceans show antagonistic interactions with other bacteria (Long and Azam, 2001), and we believe that the same is true in the ocean subsurface and overlying waters. Although bacterial predators have been commonly found in the open ocean (Long and Azam, 2001; Pineiro et al., 2007), there have not been targeted investigations to explore them in the subsea by culture-dependent and culture-independent technologies. In this investigation, our goal was to confirm the presence, diversity, and distribution of bacterial predators in the deep sea using both methodologies and, if successful in isolating Bdellovibrio and like organisms (BALOs), to determine the susceptibility of selected bacterial isolates from the subseafloor to the predators.
Among the few described predators of bacteria, the most investigated are viruses and protists; however, BALOs have received growing attention in recent decades and are the most studied among known predatory bacteria. BALOs are ubiquitously distributed in the open ocean and estuaries (Pineiro et al., 2007; Yooseph et al., 2007) and have been detected in extreme environments such as saltern ponds, the Great Salt Lake in Utah, USA (Pineiro et al., 2007), Antarctica (Jurkevitch and Davidov, 2006), and hot springs (Davidov and Jurkevitch, 2004). Their wide distribution gives us reason to believe that these predators are present in the ocean subseafloor and have an essential role in the ecology of the microbial community there.
BALOs are the only known predatory bacteria with a life cycle alternating between an extracellular free-living, highly motile phase and an intraperiplasmic growth and multiplication phase that results in the death and lysis of the prey and release of new progeny into the environment to repeat the cycle. In the extracellular free-living phase, the highly motile predators are propelled by a polar flagellum at speeds up to 160 μm/s. Based on their speed and small size (about one-fifth the size of a typical bacterium), they have been called “the world’s smallest” and fastest hunters. They attack their prey with deadly force, penetrating through the outer cell wall and into the periplasmic space. The prey cell serves as the predator’s food source and also as a growth and multiplication chamber. Although described as aerobic, BALOs have been reported to prey on biofilm bacteria under microaerophilic conditions (Kadouri and Tran, 2013) and have been found in deep ocean anoxic environments (Buttigieg and Ramette, 2014). Like Beggiotoa and some other bacteria, BALOs may inhabit the oxic/anoxic interface (Jørgensen, 1982).
The experimental design and methodologies used for detecting BALOs in samples from the subseafloor and the results are described below.
Experimental approach
Sediment samples were collected from the South China Sea during International Ocean Discovery Program (IODP) Expedition 349, South China Sea Tectonics. Here we report cored sections obtained from Site U1433 (12°55.138′N/115°02.8345′E) (Figure F1). The site is located on a structural high at the transition between the extended continental crust and the oceanic crust (Li et al., 2015). The water depth at the site was 4379.4 m, the penetration depth 188.3 m, the cored depth 188.3 m, and the recovered core length 168.79 m. The lengths of the sections sampled for predatory bacteria detection (349-U1433A-11H-3 and 19H-4) were 87 cm and 48 cm, respectively. The composition of both sections was primarily clay.
Samples were processed under anoxic conditions (glove bag with N2 + H2 atmosphere) in a cold room and stored in plastic 50 mL centrifuge tubes at 4°C or −80°C. Sediment samples were processed for the isolation of predatory bacteria, as shown in Figure F2.
Description of analytical methods
Sediment samples were collected shipboard by microbiologists (Chaunlun Zhang and Frederick Colwell) and labeled with the respective IODP sample identification information. Samples were stored at −70°C on the ship and then shipped frozen to Florida A&M University. Samples from Sections 349-U1433A-11H-3 and 19H-4 were selected for this experiment.
For the direct-plating experiment, 10 g of each sediment sample was transferred into a 50 mL sterilized disposable tube containing 20 mL of 70% sterilized artificial seawater (ASW). The sample was then suspended in the ASW by mechanical agitation for 5 min. Following agitation, the supernatant fluid was filtered sequentially through 0.8 and 0.45 µm filters (Nalgene, Rochester, NY, USA) to remove debris and larger organisms. Finally, 1 mL of the 0.45 µm filtrate was combined with 5 mL of the bacterial prey suspension (Vibrio parahaemolyticus P-5 in molten top agar) and cultured using the double agar overlay plating technique (Li and Williams, 2015). The plates were incubated at 28°C and examined for development of typical BALO-type plaques daily for up to 5 days.
BALO enrichment cultures were established by dispensing 20 mL of the 0.45 µm filtrate of the sediment samples into 500 mL Erlenmeyer flasks and amending with suspensions of the bacterial prey, V. parahaemolyticus P-5, to yield an optical density (OD) measurement of 0.7 at 600 nm. The enrichment microcosm flasks were shaken at room temperature for 120 h. A 1 mL aliquot of the enrichment samples was removed and plated using the double agar overlay technique.
Total DNA from the deep-sea sediment and control samples were extracted using the PowerSoil DNA isolation kit according to the manufacturer’s protocol. Control samples were water and sediment samples collected from the Apalachicola Bay in northwest Florida, USA.
Results and discussion
BALOs were not detected by either of the culture-based methods. Following examination of culture plates, there was no visual detection of BALO plaque formation as would be expected if the predatory bacteria were present. Likewise, the enrichment liquid cultures did not yield BALO isolates when plated onto double-layered agar plates with prey bacteria.
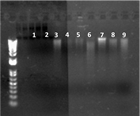
No genomic DNA was extracted from sediment samples taken from Sections 349-U1433-11H-3 and 19H-4. As shown in Figure F3, no DNA bands were observed in Lanes 1 and 2, but bands were detected in control samples (Figure F3, Lanes 3 through 9). Although BALOs have been detected by culture-independent methods from deep ocean sediments, to our knowledge, this preliminary and limited study was the first effort to recover BALOs from subseafloor samples using both culture-dependent and culture-independent approaches. More extensive studies are needed before ruling out the presence of these predators in the subseafloor environment.
Acknowledgments
This research used samples and/or data provided by International Ocean Discovery Program (IODP) Expedition 349. Funding for this research was provided by the National Institute of Minority Health and Health Disparities of the National Institutes of Health through grant Numbers 8G12MD007582-28 and 1P20MD006738-01.
References
Buttigieg, P.L., and Ramette, A., 2014. Biogeographic patterns of bacterial microdiversity in Arctic deep-sea sediments (HAUSGARTEN, Fram Strait). Frontiers in Microbiology, 5: 660. https://doi.org/10.3389/fmicb.2014.00660
Davidov, Y., and Jurkevitch, E., 2004. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax–Peredibacter clade as Bacteriovoracaceae fam. nov. International Journal of Systematic and Evolutionary Microbiology, 54(5):1439–1452. https://doi.org/10.1099/ijs.0.02978-0
Hagström, Å., Azam, F., Andersson, A., Wikner, J., and Rassoulzadegan, F., 1988. Microbial loop in an oligotrophic pelagic marine ecosystem: possible roles of cyanobacteria and nanoflagellates in the organic fluxes. Marine Ecology Progress Series, 49:171–178. https://pdfs.semanticscholar.org/f4f1/fac22729040c54239314e657b19d47f7775b.pdf
Jørgensen, B.B., 1982. Ecology of the bacteria of the sulphur cycle with special reference to anoxic–oxic interface environments. Philosophical Transactions of the Royal Society, B: Biological Sciences, 298(1093):543–561. https://doi.org/10.1098/rstb.1982.0096
Jørgensen, B.B., and Boetius, A., 2007. Feast and famine—microbial life in the deep-sea bed. Nature Reviews Microbiology, 5:770–781. https://doi.org/10.1038/nrmicro1745
Jurkevitch, E. and Davidov, Y., 2006. Phylogenetic diversity and evolution of predatory prokaryotes. In Jurkevitch, E. (Ed.), Biology, Ecology and Evolution (Volume 4): Predatory Prokaryotes: Berlin (Springer), 11–56. https://doi.org/10.1007/7171_052
Kadouri, D.E., and Tran, A., 2013. Measurement of predation and biofilm formation under different ambient oxygen conditions using a simple gasbag-based system. Applied and Environmental Microbiology, 79(17):5264–5271. https://doi.org/10.1128/AEM.01193-13
Li, C.-F., Lin, J., Kulhanek, D.K., Williams, T., Bao, R., Briais, A., Brown, E.A., Chen, Y., Clift, P.D., Colwell, F.S., Dadd, K.A., Ding, W., Hernández-Almeida, I., Huang, X.-L., Hyun, S., Jiang, T., Koppers, A.A.P., Li, Q., Liu, C., Liu, Q., Liu, Z., Nagai, R.H., Peleo-Alampay, A., Su, X., Sun, Z., Tejada, M.L.G., Trinh, H.S., Yeh, Y.-C., Zhang, C., Zhang, F., Zhang, G.-L., and Zhao, X., 2015. Expedition 349 summary. In Li, C.-F., Lin, J., Kulhanek, D.K., and the Expedition 349 Scientists, South China Sea Tectonics. Proceedings of the International Ocean Discovery Program, 349: College Station, TX (International Ocean Discovery Program). https://doi.org/10.14379/iodp.proc.349.101.2015
Li, N., and Williams, H.N., 2015. 454 pyrosequencing reveals diversity of Bdellovibrio and like organisms in fresh and salt water. Antonie van Leeuwenhoek, 107(1):305–311. https://doi.org/10.1007/s10482-014-0327-9
Long, R.A., and Azam, F., 2001. Antagonistic interactions among marine pelagic bacteria. Applied and Environmental Microbiology, 67(11):4975-4983. https://doi.org/10.1128/AEM.67.11.4975-4983.2001
Marchesi, J.R., Weightman, A.J., Cragg, B.A., Parkes, R.J., and Fry, J.C., 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia margin as revealed by 16S rRNA molecular analysis. FEMS Microbiology Ecology, 34(3):221–228. https://doi.org/10.1111/j.1574-6941.2001.tb00773.x
Pineiro, S.A., Stine, O.C., Chauhan, A., Steyert, S.R., Smith, R., and Williams, H.N., 2007. Global survey of diversity among environmental saltwater Bacteriovoracaceae. Environmental Microbiology, 9(10):2441–2450. https://doi.org/10.1111/j.1462-2920.2007.01362.x
Yooseph, S., Sutton, G., Rusch, D.B., Halpern, A.L., Williamson, S.J., Remington, K., Eisen, J.A., et al., 2007. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biology, 5(3):e16. https://doi.org/10.1371/journal.pbio.0050016
1 Williams, H.N., Li, N., and the Expedition 349 Scientists, 2018. Data report: exploring the presence of Bdellovibrio and like organisms in deep-sea sediment by culture-independent and culture-dependent methods. In Li, C.-F., Lin, J., Kulhanek, D.K., and the Expedition 349 Scientists, South China Sea Tectonics. Proceedings of the International Ocean Discovery Program, 349: College Station, TX (International Ocean Discovery Program). https://doi.org/10.14379/iodp.proc.349.202.2018
2 School of the Environment, Florida A&M University, USA. henryneal.williams@famu.edu
3 Key Laboratory of Environment Change and Resources Use in Beibu Gulf (Guangxi Teachers Education University), Ministry of Education, China.
4 Expedition 349 Scientists’ addresses.
This work is distributed under the Creative Commons Attribution 4.0 International (CC BY 4.0) license.