Teske, A., Lizarralde, D., Höfig, T.W., and the Expedition 385 Scientists
Proceedings of the International Ocean Discovery Program Volume 385
publications.iodp.org
https://doi.org/10.14379/iodp.proc.385.204.2024
Data report: the DNA event horizon in the Guaymas Basin subsurface biosphere: technical advances and redefined limits in bulk extractions of nucleic acids from deep marine sediments, IODP Expedition 3851
Gustavo A. Ramírez,2 Paraskevi Mara,3 David Beaudoin,3 Diana Bojanova,4 John E. Hinkle,5 Brewster Kingham,6 Virginia P. Edgcomb,4 Yuki Morono,4 and Andreas Teske4
1 Ramírez, G.A., Mara, P., Beaudoin, D., Bojanova, D., Hinkle, J.E., Kingham, B., Edgcomb, V.P., Morono, Y., and Teske, A., 2024. Data report: the DNA event horizon in the Guaymas Basin subsurface biosphere: technical advances and redefined limits in bulk extractions of nucleic acids from deep marine sediments, IODP Expedition 385. In Teske, A., Lizarralde, D., Höfig, T.W., and the Expedition 385 Scientists, Guaymas Basin Tectonics and Biosphere. Proceedings of the International Ocean Discovery Program, 385: College Station, TX (International Ocean Discovery Program).
https://doi.org/10.14379/iodp.proc.385.204.2024
2 California State University Los Angeles, USA
3 Woods Hole Oceanographic Institution, USA
4 Expedition 385 Scientists’ affiliations. Correspondence author: teske@email.unc.edu
5 University of North Carolina at Chapel Hill, USA.
Abstract
We compiled DNA and RNA isolation protocols for sediment bulk extraction and their yields from Guaymas Basin subsurface sediments recovered during International Ocean Discovery Program Expedition 385 and evaluated their sensitivity for metagenomic and amplicon analyses of subsurface microbial communities. Guaymas Basin sediments present a challenge for DNA and RNA recovery due to high concentrations of hydrocarbons, steep thermal gradients, and rapidly declining cell numbers downhole. Metagenomic library construction and sequencing was possible from as little as 0.2 to 0.5 ng DNA/cm3 sediment; polymerase chain reaction (PCR) amplification of 16S rRNA genes required in most cases approximately 1–2 ng DNA/cm3 sediment. At in situ temperatures of 50° to 60°C, decreasing DNA recovery leads to increasingly uncertain hit or miss outcomes and failures for metagenomic and amplicon analyses. DNA concentration profiles show that, even before these hot temperatures are reached, relatively moderate temperatures (near 40°C) have a major effect on microbial abundance and DNA yield. Comparison with cell count profiles shows that hydrothermal influence reduces downhole cell densities by multiple orders of magnitude compared to nonhydrothermal sediments. This effect is also visible at relatively moderate temperatures. RNA recovery is highly sensitive to downhole increasing temperatures and decreasing cell numbers, and was most efficient for microbial communities in cool, relatively shallow subsurface sediments.
1. Introduction
Cultivation-independent sequencing studies of microbial communities in the deep subsurface require extraction of nucleic acids in sufficient quantity and quality for subsequent high-throughput sequencing. Here we examine the outcome of DNA and RNA extractions for deep subsurface sediments from Guaymas Basin, a sedimented, hydrothermally active spreading center with steep thermal gradients and high heat flow in the axial troughs and flanking regions (Neumann et al., 2023). This study reports on nucleic acid extractions from Guaymas Basin, which have been difficult due to their high content of organic material, including hydrocarbons that can interfere with the activities of enzymes and other reagents in extraction protocols (Mara et al., 2023b). The observations we make about downhole DNA and RNA recovery will not only be useful for the community of scientists interested in Guaymas Basin but also for the broader community of scientists who study the sedimented subsurface biosphere.
Eight different sites with contrasting thermal and geochemical regimes were drilled during International Ocean Discovery Program (IODP) Expedition 385 (Teske et al., 2021a). The locations of these drill sites generally followed a northwest-to-southeast transect across the northern Guaymas Basin flanking regions and the axial trough. Two neighboring sites (U1545 and U1546) on the northwestern end of Guaymas Basin (Teske et al., 2021b; Teske et al., 2021c) essentially differ because of the presence of a massive, thermally equilibrated sill between 350 and 430 meters below seafloor (mbsf) at Site U1546 (Lizarralde et al., 2023). Two drill sites (U1547 and U1548) target the hydrothermally active Ringvent area, approximately 28 km northwest of the spreading center (Teske et al., 2019), where a shallow, recently emplaced hot sill creates steep thermal gradients and drives hydrothermal circulation (Teske et al., 2021d). Site U1549 (Teske et al., 2021e) explores the periphery of an off-axis methane cold seep, Octopus Mound, located ~9.5 km northwest of the northern axial graben (Teske et al., 2021f). Of all these sites, the two Ringvent sites have the steepest thermal gradients (between 506° and 958°C/km) and the highest heat flow values (between 516 and 929 mW/m2) (Neumann et al., 2023). This distinct thermal regime is caused by a recently emplaced, shallow sill intrusion that is driving local hydrothermal circulation (Teske et al., 2019). Initial shipboard cell counts performed on the research vessel (R/V) JOIDES Resolution indicate rapidly decreasing cell numbers with depth at all sites, starting above 109 cells/cm3 in surficial sediments but decreasing toward 106 cells/cm3 and lower (Teske et al., 2021a). The downhole decrease in cell numbers is steeper at Ringvent compared to the other sites; values near 106 cells/cm3 are reached around 60 mbsf at Ringvent, whereas comparable cell densities at the northwestern Sites U1545 and U1546 are reached around 150 mbsf (Teske et al., 2021a). In first approximation, this steep downhole decline in cell densities appears to be linked to the strong thermal gradients in the Guaymas Basin sedimentary subsurface. Rapidly decreasing downhole cell densities create special challenges for DNA and RNA extraction due to declining biomass with depth.
Here we evaluate DNA and RNA extraction methods that proved workable for these deep subsurface sediments. We report recovery of DNA (Tables T1, T2) and RNA (Table T3) obtained by these protocols, and we discuss downhole trends in microbial cell density in Guaymas Basin sediment cores (postexpedition onshore counts; Table T2) within the context of extraction yields. We provide qualified depth estimates when, based on current technical limitations, the recovery of DNA and RNA becomes insufficient to support microbial community analyses by PCR and metagenomic and metatranscriptomic sequencing. For DNA, we term these limits the DNA event horizon and suggest strategies to gradually extend these thresholds. For the Guaymas sample set, DNA recovery extends deeper into subsurface sediments than RNA recovery, which required additional washing steps to remove inhibitors. Because of the different extraction procedures for DNA versus RNA and apparent sensitivity limits, the ranges of DNA and RNA recovery in the deep subsurface are not the same; therefore, we cannot identify a combined DNA/RNA event horizon at this time. However, this RNA extraction procedure should provide a basis for further development.
2. Materials and methods
2.1. DNA extraction
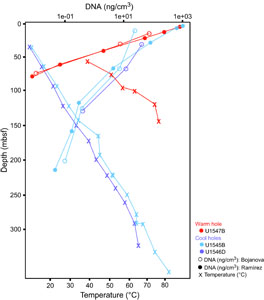
DNA was extracted from selected core samples using a FastDNA SPIN Kit for Soil (MP Biomedicals). For each sample listed in Table T1, three parallel (triplicate) DNA extractions were performed. The three extracts were pooled together and concentrated as discussed below. The samples listed in Table T2 were extracted once (single extraction). Otherwise, sediments were processed following the manufacturer’s protocol with homogenization modifications as described previously (Ramírez et al., 2018). We note that some DNA extraction sets (Figure F1; Table T1) started with volumetrically defined wet sediment samples of 0.5 cm3 each, whereas others started with weighed wet sediment samples of 0.5 g each (Figure F2; Table T2). These concentrations can be converted into each other to an approximate degree by using a conversion factor of 1.7 g/cm3, which is the average wet bulk density of an extensive set of IODP sediments (Tenzer and Gladkikh, 2014). The extraction procedure tolerates a sediment input of 0.5 cm3, which weighs more than 0.5 g. Procedures should be kept internally consistent for each sample set, and the extraction vials have to be balanced during the centrifugation steps.
Briefly, each sediment sample was homogenized twice (in contrast to the manufacturers’ suggestion of a single homogenization step) in Lysing Matrix E tubes for 40 s at a speed of 5.5 m/s, using the MP biomedicals bench top homogenizer equipped with 2 mL tube adaptors. Between the two homogenization rounds the samples were placed on ice for 2 min. After the second homogenization, the samples were centrifuged at a relative centrifugal force of 14,000 times gravity (× g) for 5 min following the manufacturer’s suggestion. In the next step, which was a second modification from the standard protocol (applied only to DNA extractions of Table T1), the supernatant and the top layer of the pellet was transferred to a clean 2 mL tube, where proteins were precipitated by the addition of the protein precipitation solution provided in the extraction kit. The rest of the extraction protocol followed the manufacturer’s recommendations. When parallel extractions were performed, the extracts were pooled and concentrated using Amicon Ultra 0.5 mL 30K Centrifugal Filters (Millipore Sigma). The DNA concentration was fluorometrically measured using the high sensitivity (HS) double-stranded (ds) DNA Qubit assays (Qubit dsDNA HS and BR Assay Kits). A kit control extraction (extraction with no sediment added; blank, or witness, extraction) was included to account for any potential kit and handling contaminants. For extractions from deeper sediments where triplicate sample pooling did not result in quantifiable amounts of DNA, we performed 10 parallel extractions that were subsequently pooled and concentrated using the Amicon filters as described above.
2.1.1. Protocol A
Protocol A is used for high biomass samples with expected cell densities ≥ 106 cells/cm3. Three parallel extractions are done per sample, including the blank, or witness, extractions.
- Add up to 0.5 cm3 sediment or 0.5 g sediment to a Lysing Matrix E tube.
- Add 978 µL Sodium Phosphate Buffer to sample in Lysing Matrix E tube.
- Add 122 µL of modified Tyrode’s buffer (MT buffer) solution.
- Homogenize in the FastPrep homogenizer or equivalent bead-beating homogenizer for 40 s twice at speed setting of 5.5 m/s with 2 min rest periods on ice.
- Centrifuge at 14,000 × g for 5 min to pellet debris.
- Transfer supernatant to a clean 2 mL tube. (Note: At this step, a modification of the manufacturer’s protocol is recommended. Carefully aspirate a few [50–100 µL max] of the top part of the pellet as well. Avoiding the pellet leads to significantly lower DNA yields, meaning that the positive charges on pelleted particles sequester DNA. After bead-beating, DNA is extracellular and fully exposes its negatively charged phosphodiester backbone. This trapped DNA is subsequently released into Sodium Phosphate Buffer.)
- Add 250 µL protein precipitating solution and mix by shaking by hand 10 times.
- Centrifuge at 14,000 × g for 15 min to precipitate the pellet. (Note: because of the intentional pellet carryover from step 6, 15 min are allowed for protein precipitation.) Transfer the supernatant into a clean 15 mL Falcon tube.
- Resuspend the Binding Matrix suspension and add 1 mL to the supernatant in the 15 mL Falcon tube.
- Place on rotator or invert by hand for 2 min to allow binding of DNA. Place tube in rack for 3 min to allow settling of the Binding Matrix.
- Remove and discard 500 µL of supernatant, being careful to avoid settled Binding Matrix.
- Resuspend Binding Matrix in the remaining amount of supernatant. Transfer approximately 600 µL of the mixture to a SPIN Filter and centrifuge at 14,000 × g for 1 min. Empty the catch tube, add 600 µL of the mixture and centrifuge again at 14,000 × g for 1 min. Repeat process until all mixture passes through the SPIN filter.
- Add 500 µL prepared SEWS-M (Nucleic Acid Wash Solution provided by the kit) and gently resuspend the Binding Matrix pellet using the force of the liquid from the pipette tip. Ensure that absolute ethanol has been added to the concentrated SEWS-M stock solution as required by the manufacturer.
- Centrifuge at 14,000 × g for 1 min. Empty the catch tube.
- Centrifuge again at 14,000 × g for 2 min to “dry” the Binding Matrix from residual wash solution. Discard the catch tube and replace with a new, clean catch tube.
- Air dry the SPIN Filter for 5 min at room temperature.
- Gently resuspend the Binding Matrix (collected inside the SPIN filter) in 50 µL of DNase/Pyrogen-free water (DES). (Note: not 30 µL of DES, as in manufacturer’s protocol.)
- Centrifuge at 14,000 × g for 1 min to elute DNA from the Binding Matrix into the clean catch tube. Discard the SPIN filter. This will result in ~50 µL of DNA in the catch tube. (Note: use the smallest possible volume [1 µL] for Qubit fluorometric quantification to save DNA.) Store all extracts or single pooled extracts at −80°C.
2.1.2. Protocol B
Protocol B is used for low biomass samples with expected cell densities are < 106 cells/cm3. A total of 10 parallel extractions were performed per sample and include a batch of blank, or witness, extractions. Steps 1–18 are the same as in Protocol A, performed 10 times in parallel.
- Potential stopping point or break: store all extracts or single pooled extracts at −80°C now if necessary.
- Assuming that yields are not sufficient (which is nearly always the case if cell concentrations are expected < 106 cells/cm3 in the sample), transfer the full combined volume of all 10 individual extractions (~500 µL) to an Amicon Ultra 0.5 mL 30K filter for concentrating the DNA, following the manufacturer’s instructions for sample concentration (steps 1–7), omitting pre-rinsing and desalting.
2.1.3. Protocol C
Protocol C improves the DNA yield by adding a lysozyme and proteinase K step before performing DNA with FastDNA SPIN kit for soil (MP Biomedical). Protocol C was tested with a shallow subsurface sediment from 0.8 mbsf (Hole U1546B), and a method control. One 0.5 g sediment sample was pretreated with lysozyme and proteinase K, and the other sample was not. The sample with the lysozyme and the proteinase K pretreatment before performing extraction Protocol A gave a higher DNA yield when compared to the nonpretreated sediment (0.8 ng/µL versus 0.2 ng/µL fluorometrically quantified using a Qubit dsDNA HS assay kit). The method control did not yield DNA. The applied heating steps (37° and 55°C for 45 and 20 min, respectively) improved the DNA extraction efficiency but also seemed to remove inhibitory compounds that interfere with DNA extraction (e.g., humic acids and hydrocarbons). The protocol could be scaled up and tested further in triplicate or tenfold extractions as described in Protocol B.
- Prepare 50 mM lysozyme solution by diluting 2 mg of lysozyme in 40 µL of Sodium Phosphate Buffer provided by the FastDNA SPIN kit for soil (MP Biomedical).
- Prepare 10 mM proteinase K solution by diluting 1 mg of proteinase K in 100 µL of Sodium Phosphate Buffer provided by the FastDNA SPIN kit for soil (MP Biomedical).
- Add up to 500 mg of sediment sample to a Lysing Matrix E tube.
- Add 800 µL Sodium Phosphate Buffer to sample in Lysing Matrix E tube and the 40 µL of the lysozyme solution.
- Incubate at 37°C rotating for 45 min. Incubation time can increase up to 1 h.
- After lysozyme treatment, add 100 µL proteinase K solution and 100 µL of 20% sodium dodecyl sulfate (SDS) (weight-to-volume ratio [w/v]) to the sample in the Lysing Matrix E tube. The addition of 20% SDS can be replaced by 120 µL MT buffer provided by the kit. (Note: SDS concentration in MT buffer is undisclosed, but SDS is known to stimulate the activity of proteinase K even at low concentrations of 0.5%–2% w/v [Hilz et al., 1975].) Replacement of 20% SDS with 120 µL MT was also tested and worked.
- Incubate at 55°C rotating for 20 min. Incubation time can increase up to 40 min.
- Place the Lysing Matrix E tube with the sample in the FastPrep homogenizer for 40 s at speed setting of 6 m/s.
- Continue with the DNA extraction Protocol A from Steps 5 through 18.
- Assuming that DNA yields are not sufficient, continue with Steps 19 and 20 described in Protocol B.
2.1.4. Other methods
We also tested DNA isolation with the Qiagen DNAeasy PowerSoil Pro kit but obtained an order of magnitude lower DNA yields compared to the FastDNA kit. Three samples (385-U1545C-4H-3, 40/60 cm; 8H-3, 40/60 cm; and 385-U1547B-3H-3, 10/30 cm) yielded 2.876, 0.684, and 4.648 ng DNA/g sediment with the Qiagen kit and 164.25, 8.66, and 104.4 ng/g sediment with the FastDNA kit, respectively. Likewise, DNA extractions using higher sediment volumes (up to 10 g) with Qiagen DNeasy PowerMax Soil also led to low DNA yields (similar ranges as those reported for Qiagen DNAeasy PowerSoil Pro kit tests). Because higher volumes of sediment also increase the concentrations of inhibitory compounds found in Guaymas subsurface sediments, our teams concluded that small-scale (0.5 cm3 or 0.5 g) sediment extractions offer a more favorable balance of DNA yield and inhibitor accumulation. These results confirmed the decision of all parties to use the modified FastDNA procedure as outlined above in Protocols A or B.
2.2. RNA extraction
To demonstrate microbial activity and associated gene expression, RNA extraction, sequencing and metatranscriptomic analyses are essential. Prior protocols for deep subsurface total RNA extraction from our laboratories involved bead-beating, organic extraction, and ethanol precipitation (Sørensen and Teske, 2006; Biddle et al., 2006) or used soil RNA extraction kits that excel for samples that have high humic content, including sediments (Edgcomb et al., 2011; Orsi et al., 2013a, 2013b). However, for over a year of effort, Guaymas Basin subsurface sediments maintained at −80°C defeated all attempts at RNA extraction using various methods, including the original RNA isolation protocols published by Chomczynski and Sacchi (1987). Aside from the different RNA isolation protocols, we performed different sediment pretreatments that have been shown to increase cell extraction efficiency from deep subsurface sediments (Kallmeyer et al., 2008). However, even with these pretreatments (e.g., acidification of sediments for 10 min with 0.43 M sodium acetate to dissolve carbonates), RNA could not be isolated. The steps that eventually led to successful RNA extraction involved initially washing sediment samples twice with absolute ethanol (200 proof; purity ≥ 99.5%; Thermo Scientific Chemicals), followed by one wash with diethyl pyrocarbonate (DEPC)-treated, RNAse-free water (Fisher BioReagents) before extracting RNA. This procedure appears to remove sufficient hydrocarbons and other inhibitory elements present in Guaymas sediments. Similar washing procedures have been tested for other hydrocarbon-rich sediments (Lappé and Kallmeyer, 2011). Without ethanol and DEPC water washes, all attempts resulted in low or zero RNA yield. At the time of writing this manuscript, metatranscriptomic analyses of Guaymas Basin subsurface sediments (Mara et al., 2023b) depended on including these washing procedures. We also note that washing steps might be applied to future DNA extraction protocols to remove inhibitors that accumulate when extraction volumes are scaled up.
In brief, 10–15 g of frozen Guaymas Basin sediments were transferred into UV-sterilized 50 mL Falcon tubes (RNAse/DNAse-free) using clean, autoclaved and ethanol-washed metallic spatulas. Each tube received an equal volume of absolute ethanol and was shaken manually for 2 min followed by 30 s of vortexing at full speed to create a slurry. Samples were transferred to an Eppendorf centrifuge (5810R) and were centrifuged at room temperature for 2 min at 2000 rpm. The supernatant was decanted, and the ethanol wash was repeated. After decanting the supernatant of the second ethanol wash, an equal volume of DEPC water was added to each sample. Samples were manually shaken and vortexed as before to create slurry and were transferred to the Eppendorf centrifuge (5810R), where they were centrifuged at room temperature for 2 min at 2000 rpm. The supernatant was decanted, and each sediment sample was immediately divided into three bead-containing 15 mL Falcon tubes provided by the PowerSoil Total RNA Isolation Kit (Qiagen). RNA was extracted as suggested by the manufacturer with the modification that the RNA extracted from the three aliquots was pooled into one RNA collection column and eluted at 30 µL final volume. All RNA extractions were performed in a UV-sterilized clean hood (two UV cycles of 15 min each) that was installed with HEPA filters. Surfaces inside the hood and pipettes were sterilized with RNase AWAY (Thermo Scientific) before every RNA extraction and in between extraction steps. Trace DNA contaminants were removed from RNA extracts using TURBO DNase (Thermo Fisher Scientific) and the manufacturer’s protocol. Carryover DNA removal from the RNA extracts was confirmed with PCR reactions using primers for the small ribosomal subunit of 16S rRNA gene (BACT1369F: 5’CGGTGAATACGTTCYCGG3’ and PROK1541R: 5’AAGGAGGTGATCCRGCCGCA 3’; Suzuki et al., 2000). Each 25 µL PCR reaction was prepared using GoTaq G2 Flexi DNA Polymerase (Promega) and contained 0.5 U/µL GoTaq G2 Flexi DNA Polymerase, 1X Colorless GoTaq Flexi Buffer, 2.5 mM MgCl 2 , 0.4 mM dNTP Mix (Promega), 4 µM of each primer (final concentrations), and DEPC water. Further, to confirm the absence of DNA contamination due to handling and PCR reagents, all PCR experiments included negative controls (blanks) where no DNA was added. PCR amplifications were performed in an Eppendorf Mastercycler Pro S Vapoprotect (Model 6321) thermocycler with following conditions: 94°C for 5 min followed by 35 cycles of 94° (30 s), 55° (30 s), and 72°C (45 s). The PCR products were run in 2% agarose gels (Low-EEO/Multi-Purpose/Molecular Biology Grade Fisher BioReagents) to confirm the absence of DNA amplicons. RNA quantification (ng/µL) was performed using Qubit RNA HS, broad range (BR), and extended range (XR) Assay Kits (Invitrogen). Because of the essential sediment washing steps with ethanol and DEPC-treated water and the small volume of final RNA extractions (30 µL), the RNA integrity of the extracted RNA was not estimated. The washes with absolute ethanol can enhance the already high susceptibility of RNA to degradation at room temperature. Therefore, we selected to preserve the small volume of our RNA extracts for other mandatory steps involved in cDNA library preparation. These steps included quantifying the total RNA concentration before the cDNA library preparation, performing PCR reactions to confirm the absence of carryover DNA in the RNA extracts, and maintaining the necessary initial RNA volume (up to 10 µL), which is required as a template for the synthesis of the single/first cDNA strand.
Amplified cDNAs from the DNA-free RNA extracts were prepared using the Ovation RNA-Seq System V2 (Tecan) following the manufacturer’s suggestions. All steps from RNA extraction through cDNA preparation were completed on the same day to avoid freeze/thaw cycles that might damage the integrity of RNA strands. cDNAs were submitted to the Georgia Genomics and Bioinformatics Core facility for library preparation and sequencing using NextSeq 500 PE 150 High Output (Illumina). Sequencing of the cDNA library prepared from the control sample (laboratory reagent control) was unsuccessful because it failed to generate any sequences that met the minimum length criterion of 300–400 base pairs.
Although this RNA extraction and transcription protocol ran into downhole detection limits (Table T3), we note that every living subsurface cell with intact genomic DNA requires at least a minimum level of gene expression to survive and function, and we therefore regard RNA extraction and detection limits foremost as methodological issues, not as evidence of fundamental constraints on life.
2.3. Results
2.3.1. Downhole trends in DNA yield
To obtain DNA for metagenomic analyses, Protocols A and B were applied to DNA extractions from Hole U1545B, U1545C, U1546D, and U1547B sediment samples, starting with 0.5 cm3 wet sediment samples. DNA yields are tabulated in Table T1 and plotted in Figure F1. The yield of DNA extractions decreases exponentially with sediment depth and temperature. In near-surface sediments, DNA yields are high, around 1000–1500 ng DNA or 1–1.5 µg DNA/cm3 wet sediment. DNA yields decrease toward the limit of reliable detection and quantification by Qubit (below 0.1 ng/cm3) within 75 mbsf in Hole U1547B at Ringvent, and within about 200 mbsf in Hole U1545B in the northwestern Guaymas Basin. The steep thermal gradient at Ringvent means that a depth of 75 mbsf corresponds to in situ temperatures near 50°–55°C, whereas the cooler sediments in the northwestern Guaymas Basin reach these temperatures only near 200 mbsf (Figure F1; Table T1). The temperature-related differences in DNA recovery impact the depth horizons for successful metagenomic sequencing, which generally requires nanogram amounts of DNA. For example, generating Illumina libraries from low-biomass samples requires a previously reported minimum of 3.65 ng DNA (Jiang et al., 2015).
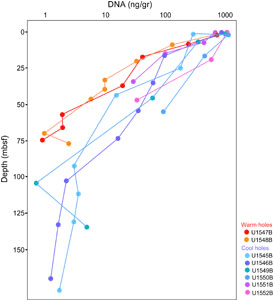
Independently, Protocols A and B were applied to extract DNA for another sequencing project from all sites; here extractions started with 0.5 g wet weight samples (Figure F2; Table T2). The DNA concentrations show a similar decline over several orders of magnitude as observed for the previous set of extractions. At the northwestern Sites U1545 and U1546, DNA concentrations decline over almost three orders of magnitude from the microgram to the nanogram range per gram sediment within the upper 170–190 m (40°–45°C). DNA yields decline over three orders of magnitude at Ringvent Sites U1547 and U1548, and this sharp decline occurs within the upper 70–80 m (50°–60°C); some uncertainty for the deeper samples is not resolved because DNA concentrations dip below the 0.1 ng limit of Qubit detection (Table T2). At the cold seep–affiliated Site U1549 and the axial graben Site U1550, DNA concentrations decline by three orders of magnitude within approximately 140 mbsf (25°–30°C). The shorter DNA gradients of southeastern Site U1551 and northern seep Site U1552 drop by more than one or two orders of magnitude over depth ranges of 35 and 45 m, respectively, and remain entirely within cool temperatures of ≤16°C (Table T2).
2.3.2. Determination of the DNA event horizon
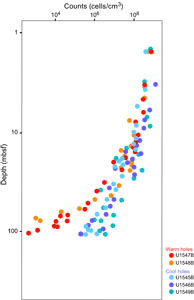
Empirically, the current limit for contaminant-free DNA that is consistently suitable for metagenomic analysis of Guaymas Basin sediments is around 0.2–0.5 ng extracted DNA/cm3 wet sediment (Table T1). Lower DNA concentrations near 0.1 ng/cm3 may still yield metagenomes, but contaminants hypothesized to emerge from the reagent’s kitome (Salter et al., 2014) increasingly appear once DNA concentrations decrease to 0.1 ng extracted DNA/cm3. Assuming a cellular DNA content of approximately 1–2 fg (10−15 g) DNA/cell (Bakken and Olsen, 1989; Button and Robertson, 2001), 0.2–0.5 ng (10−9 g) DNA is the equivalent of 0.2 × 106 to 0.5 × 106 cells. Therefore, given the complexity of our sample set, DNA of sufficient quantity and quality for metagenomic library construction and sequencing can be extracted from sediments with cell densities of approximately 0.2 × 106 to 0.5 × 106 cells/cm3, which is more than three orders of magnitude below near-surface cell densities of 109 cells/cm3 sediment. Such a decline in cell numbers is broadly consistent with parallel cell counts (Figure F3). From the perspective of obtaining sufficient DNA for metagenomes, this DNA quantity is the current DNA event horizon for this complex environment given our methodological approaches. Although this metagenomic DNA limit lags much behind the greater sensitivity of cell counts, it has improved considerably compared to earlier attempts to obtain subsurface metagenomes from nonamplified DNA extracts, which succeeded only for shallow subsurface sediments (Biddle et al., 2008, 2011). We also note that the successful preparation of metagenomic libraries from small amounts of DNA is crucial, requiring in some cases extra efforts at library preparation, and in this regard not all sequencing facilities may be equally capable. The participants of this study had their metagenomic libraries prepared at the DNA Sequencing and Genotyping Center of the University of Delaware (USA).
The DNA event horizon is slightly different for targeted PCR amplification of specific genes, and depends not only on drill site and depth but also on the amplification target (Table T2). PCR allows for the amplification of targeted gene sequences within dilute DNA extracts, increasing the sensitivity of detection. DNA extracts from all drill sites were used for PCR amplification of bacterial and archaeal 16S rRNA gene segments using primer combinations of 515F-Y (Parada et al., 2016) and 926R (Quince et al., 2011), partial archaeal 16S rRNA genes using primer combination 25F and 806R (Mara et al., 2023a), and mcrA genes (Hinkle et al., 2023) using primer combinations mcrIRD and mcrANME1 (Lever and Teske, 2015). In contrast to the metagenomic extractions in Table T1, these DNA extractions for PCR were performed in single samples rather than in triplicate, and the sample size (0.5 g) was based on wet weight, not volume. A total of 10 replicate extractions (Protocol B) were performed for a second PCR attempt if the initial PCR did not work (Table T2).
In our PCR survey, DNA concentrations near 1–2 ng DNA/g wet sediment were generally required for positive PCR outcomes (Table T2), whereas DNA samples at lower concentrations—which includes the previously defined metagenome sequencing limit (<0.2 to 0.5 ng DNA/cm or below 0.34–0.85 ng DNA/g wet weight sediment, assuming a conversion factor of 1.7)—did not produce PCR amplicons of 16S rRNA genes or other genes. The only exceptions were low-DNA extracts from deep, hot Ringvent samples (Hole U1547B) that yielded archaeal PCR products (Table T2). This surprising result of slightly higher DNA concentration requirements for PCR than for metagenomic sequencing may reflect the fact that metagenomic library preparation does not select for specific genes, which necessarily account only for a very small proportion of the DNA pool. In contrast, PCR assays for 16S rRNA genes and functional genes pick out specific genes by design, like the proverbial needle in the haystack. Interestingly, PCR amplification of an 800 base pair (bp) archaeal 16S rRNA gene segment was more consistently successful than amplification of shorter (300 bp) 16S rRNA gene segments using general prokaryotic (bacterial and archaeal) primers. Archaeal 800 bp amplicons were consistently recovered to approximately 75 mbsf in Ringvent Holes U1547B and U1548B and 170 mbsf in the northwestern Holes U1545B and U1546B (Table T2). PCR amplification of mcrA genes with the ANME1-targeted primers gave positive results generally near and within methane-sulfate interfaces (Hinkle et al., 2023). Amplification of mcrA gene for methanogens worked only for a few samples, which is consistent with the apparent rarity of methanogens in the Guaymas Basin subsurface (Bojanova et al., 2023). The robustness of archaeal 16S rRNA gene-directed PCR assays over a wide spectrum of DNA concentrations and depths may result from several factors: the absence of archaea from common laboratory and kit contaminants (Salter et al., 2014), the highly conserved archaea-specific primer sites that contrast with the more ambiguous primer regions used for general prokaryotic (bacterial and archaeal) 16S rRNA gene surveys, and the downhole increasing relative proportion of archaea in the microbiome of hydrothermal sediments (Ramírez et al., 2021; Lagostina et al., 2021).
DNA yields in the Guaymas subsurface decline by three orders of magnitude by 50–100 mbsf (Figures F1, F2). We see declining DNA yields already in sediments with warm in situ temperatures of 35°–40°C (40–50 mbsf at Ringvent sites and 130–150 mbsf at Site U1545), which is far below the temperature range for hyperthermophilic vent archaea (80°C and higher), indicating that the survival of most subsurface microorganisms is already noticeably reduced at those depths. The metagenomic analysis showed that mesophilic microbiota dominate in Guaymas Basin sediments, indicating that the downhole decreases of cell abundance and DNA yields are mainly caused by environmental selection against these microbes (Mara et al. 2023a). In nonhydrothermal sediments from passive continental margins such a decline in cell numbers occurs over well above 1000 mbsf (Parkes et al., 2000, 2014), corresponding to successful DNA and RNA recovery and subsequent sequencing surveys at considerably greater depth compared to Guaymas Basin (Inagaki et al., 2006; Biddle et al., 2006; Pachiadaki et al. 2016).
Potential DNA yields extrapolated from cell counts (Figure F3) often yielded considerably lower values compared to DNA extractions, by one order of magnitude or more; these differences appear to increase with depth and suggest an increasing contribution from extracellular DNA (Table T2). However, directly comparing microscopic cell counts and DNA extractions requires caution because inherent assumptions need to be considered. Comparing them directly assumes a constant averaged cell-specific DNA amount, but genome size decreases with depth and in situ temperature (Mara et al. 2023a). Matching cell count–derived and extracted DNA amounts also requires negligible quantitative contributions of extracellular DNA to the total DNA pool in marine sediments, which is a much-debated topic and active research field (Torti et al. 2015).
We note that the depth range horizon for successful metagenomic library construction, sequencing, and recovery of metagenome-assembled genomes (MAGs) is very different from cell count limits. The Expedition 385 summary chapter includes shipboard cell counts that reach 105 to 106 cells/cm3 (Teske et al., 2021a). In general, samples with these concentrations yield sufficient DNA for metagenomic sequencing and MAG analysis. Postexpedition counts using automated image acquisition and high-throughput counts in the laboratory will be much more sensitive and will enable more precise quantification of subsurface cell density in Guaymas Basin to 102 to 103 cells/cm3 (Morono et al., 2022). Obviously, these sparser deep subsurface microbial communities also contain intracellular DNA that should in principle be amenable to sequencing. However, bridging the sensitivity gap between metagenomics (limited by DNA yield) and cell counts (limited by high-throughput microscopic image processing) remains a challenge and will require continued development of combined cell separation/DNA extraction approaches (Kallmeyer et al., 2008; Morono et al., 2013). So far, extremely deep and hot samples where cell counts and activity measurements suggest persistent microbial life remain inaccessible by sequencing (Heuer et al., 2017, 2020; Beulig et al., 2022).
2.3.3. Comparison to shallow hydrothermal sediments
Near-surface densities of 109 to 1010 cells/cm3 are found in hydrothermal sediments of Guaymas Basin. DNA concentrations in surficial hydrothermal sediments range 6 (Hinkle et al., preprint) to 10 µg/cm3 (Engelen et al., 2021). Based on the average DNA content of 1–2 fg (10−15 g) DNA/cell, these DNA concentrations translate into cell densities of 3 × 109 to 10 × 109 cells/cm3 ; these numbers match the range of epifluorescence cell count maxima in surficial sediments of Guaymas Basin that range 1 × 109 to 4 × 109 cells/cm3 (Meyer et al., 2013). Given high cell numbers and DNA concentrations, metagenomic sequencing of microbial communities in surficial sediments is not limited by DNA availability and yields highly diverse bacteria and archaeal communities with novel lineages (Dombrowski et al., 2017, 2018; Seitz et al., 2019; Eme et al., 2023).
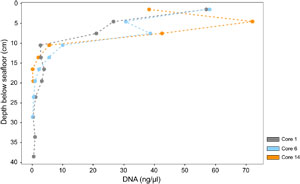
In contrast to the deep Guaymas Basin subsurface, DNA yields in surficial hot hydrothermal sediments of Guaymas Basin sampled by submersible using push cores (Alvin Dives 4869 and 4872) decrease by several orders of magnitude but on scales of centimeters instead of tens or hundreds of meters. Mutually independent studies show that these downhole decreases in DNA yield are persistent regardless of different DNA extraction protocols and quantification units (Engelen et al., 2021; Hinkle et al., preprint). DNA yields from a hot hydrothermal sediment push core declined over one order of magnitude from 10 to <1 µg DNA/g sediment within the top 3 cm, using the Qiagen power soil extraction kit and protocol (Engelen et al., 2021). DNA yields from three sediment push cores along a hydrothermal gradient declined more than two orders of magnitude from 6 µg/cm3 to <60 ng/cm3 within the top 20 cm (Figure F4; Table T4) using a manual extraction protocol (modified from Zhou et al., 1996) that combines freeze-thawing, proteinase K digest, and chloroform-isoamyl alcohol extraction (Hinkle et al., preprint). We note that these surficial hydrothermal sediments are very soupy, which minimizes the difference between volume-based and weight-based DNA yield.
This strong compression of high DNA yields and microbial populations toward the sediment/water interface is commonly ascribed to increasingly extreme temperatures and biogeochemical conditions in hydrothermally active shallow sediments (McKay et al., 2016; Teske et al., 2016; Su et al., 2023). These results show some qualitative similarities to the Expedition 385 results, insofar as hydrothermal stress factors appear to compress DNA yield and microbial cell numbers toward the upper subsurface sediments of Guaymas Basin and depress DNA yield and microbial cell numbers in the subsurface. Although the impacts of temperature stress and energy limitation apply to both shallow and deep sediments, the conditions and hydrothermal settings are certainly different. The fluidized surficial hydrothermal sediments that are permeated by pulsating, extremely hot (>80°C) and highly reducing fluids (McKay et al., 2016; Teske et al., 2016; Su et al., 2023) are distinct from consolidated deeper subsurface sediments characterized by stable thermal and geochemical gradients. Interestingly, the microbial communities of dynamic near-surface hydrothermal sediments and of hydrothermally influenced subsurface sediments are different; the former are dominated by hydrothermal vent and hot spring taxa, whereas the latter share many microbial groups with the global marine sedimentary biosphere (Lagostina et al., 2021). We speculate that microbial communities in hot, hydrothermally active surficial sediments have access to carbon sources, nutrients, and redox pairs that provide sufficient energy to cope with extreme temperature stress; in contrast, relatively moderate temperatures that are measured in IODP-drilled subsurface sediments have a disproportionally greater impact on the energy-limited microbial deep biosphere (Mara et al., 2023a, 2023b). These differences in selection factors may ultimately result in distinct microbial communities.
2.3.4. Strategies to reach beyond the DNA event horizon
Obtaining sufficient DNA from sediments with lower cell numbers requires further methodological improvements, for example separating cells from sediments before cell rupture and DNA extraction to prevent DNA sticking to hydrocarbon-coated sediment particles (Lappé and Kallmeyer, 2011). Refined methods for cell separation from sediments and subsequent concentration have been reviewed and discussed in detail in the context of cell counting (Morono, 2023) and will ultimately lead to increased sequencing sensitivity as well.
Bulk sediment DNA isolation procedures can be scaled up using multiple parallel extractions followed by pooling of the extracted DNA, for example by changing from Protocol A (1 or 3 parallel extractions per sample) to Protocol B (10 parallel extractions per sample). As a caveat, in the course of pooling and concentrating multiple samples, laboratory or reagent contaminants that are present at low levels would also be concentrated and collected. This unintended side effect may have resulted in Actinobacterial and certain Gammaproteobacterial contaminants that appear increasingly in metagenomic sequencing and MAG annotation for low-DNA samples (Mara et al., 2023a). To reduce the need for multiple parallel extractions of small volumes, sediment washing steps similar to those used for RNA isolation could be applied to DNA extraction procedures to scale up extraction volumes without accumulating inhibitor substances or process-derived contaminants in the final extracts.
With the advent of advanced DNA sequencing technologies and the laboratory methods to support them, sequencing projects targeting recalcitrant and extreme samples now have a significantly higher rate of success (i.e., Tighe et al., 2017). Reactive and corrosive compounds can co-elute with nucleic acids while implementing typical purification methods, and these compounds often negatively affect downstream procedures. Further, organisms that survive in these environments possess cellular mechanisms to protect their genomes for gene expression and reproduction. Interestingly, these evolved cellular mechanisms that protect DNA from environmental damage can make the DNA more difficult to extract and to analyze, often inhibiting downstream enzymatic reactions or requiring harsh extraction techniques that result in highly fragmented DNA (Morono et al., 2014). New methods and products have been developed that can more effectively remove protective and reactive and/or corrosive compounds. Industry vendors that have stepped into this space with products include Zymo Research (https://www.zymoresearch.com), Qiagen (https://www.qiagen.com/us), Omega Bio-Tek (https://omegabiotek.com), and others. Home brew methods that have been developed are as simple as performing serial dilutions to reduce inhibitory compounds or more complex as with newly formulated reagents and solutions. The ability to overcome these challenges has led to a surge in high quality data from previously inaccessible sample materials (Bojanova et al., 2023; Mara et al., 2023a, 2023b).
Computational methods may also expand the limits of useful metagenomic surveys beyond the currently proposed DNA event horizon despite potential contaminant loads. Taxonomy-based identification of contaminants in a metagenomic sequencing datasets may help eliminate unwanted biological information at the short-read, contiguous sequence (contigs) assembly, and/or open reading frame (protein prediction) stage. Generating large numbers of contigs that are taxonomically assigned to known contaminants can improve metagenomic assembly quality by culling the assembly input. Alternatively, independent taxonomic assignments for all proteins predicted in metagenomic contigs can serve as another contaminant culling step with the potential to increase the number of MAGs or the percentage of complete/near-complete MAGs from a given data set (https://github.com/Arkadiy-Garber/Taxonsluice). To extract information from Kyoto Encyclopedia of Genes and Genomes (KEGG) modules for incomplete bacterial MAGs, MetaPathPredict (Geller-McGrath et al., 2024) can generate predictions for the presence or absence of KEGG modules within gene annotations of bacterial MAGs; we have recently applied this approach to the Guaymas deep biosphere (Mara et al., 2023a).
To conclude, we note that further methods development can proceed using archived frozen sediment samples after the retirement of R/V JOIDES Resolution to make productive use of this pause in deep drilling. Advancing the sensitivity of sequence-based microbial community analyses toward the biotic/abiotic interface in the deep subsurface remains essential for understanding the astonishing tenacity of deep subsurface life.
3. Acknowledgments
This study was supported by National Science Foundation (NSF) Grant OCE-2046799 to V.P. Edgcomb, P. Mara, and A. Teske; NSF grant OCE-1829903 to V.P. Edgcomb, P. Mara, and A. Teske; NASA Exobiology Grant APP-0244-001 to A. Teske; NSF Grant OCE-0939564 to D. Bojanova, and JSPS KAKENHI Grants JP19H00730 and JP23H00154 to Y. Morono. IODP Expedition 385 participants were aided by IODP cruise and postexpedition support. We thank all IODP Expedition 385 scientists, technicians, drillers, and crew for making sample recovery, and by proxy, this research project possible. We gratefully acknowledge the shipboard curatorial team who kept the sediment samples and metadata organized and well cataloged. We thank Amend Laboratories for hosting G. Ramírez for the first rounds of DNA extractions before the COVID-19 pandemic shutdown. We are grateful for the constructive comments of an anonymous reviewer.
References
Bakken, L.R., and Olsen, R.A., 1989. DNA-content of soil bacteria of different cell size. Soil Biology and Biochemistry, 21(6):789–793. https://doi.org/
Beulig, F., Schubert, F., Adhikari, R.R., Glombitza, C., Heuer, V.B., Hinrichs, K.U., Homola, K.L., Inagaki, F., Jørgensen, B.B., Kallmeyer, J., Krause, S.J.E., Morono, Y., Sauvage, J., Spivack, A.J., and Treude, T., 2022. Rapid metabolism fosters microbial survival in the deep, hot subseafloor biosphere. Nature Communications, 13(1):312. https://doi.org/
Biddle, J.F., Fitz-Gibbon, S., Schuster, S.C., Brenchley, J.E., and House, C.H., 2008. Metagenomic signatures of the Peru margin subseafloor biosphere show a genetically distinct environment. Proceedings of the National Academy of Sciences of the United States of America, 105(30):10583–10588. https://doi.org/
Biddle, J.F., Lipp, J.S., Lever, M.A., Lloyd, K.G., Sorensen, K.B., Anderson, R., Fredricks, H.F., Elvert, M., Kelly, T.J., Schrag, D.P., Sogin, M.L., Brenchley, J.E., Teske, A., House, C.H., and Hinrichs, K.-U., 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proceedings of the National Academy of Sciences of the United States of America, 103(10):3846–3851. https://doi.org/
Biddle, J.F., White, J.R., Teske, A.P., and House, C.H., 2011. Metagenomics of the subsurface Brazos-Trinity Basin (IODP Site 1320); comparison with other sediment and pyrosequenced metagenomes. The ISME Journal, 5(6):1038–1047. https://doi.org/
Bojanova, D.P., De Anda, V.Y., Haghnegahdar, M.A., Teske, A.P., Ash, J.L., Young, E.D., Baker, B.J., LaRowe, D.E., and Amend, J.P., 2023. Well-hidden methanogenesis in deep, organic-rich sediments of Guaymas Basin. The ISME Journal. https://doi.org/
Button, D.K., and Robertson, B.R., 2001. Determination of DNA content of aquatic bacteria by flow cytometry. Applied and Environmental Microbiology, 67(4):1636–1645. https://doi.org/
Chomczynski, P., and Sacchi, N., 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry, 162(1):156-159. https://doi.org/
Dombrowski, N., Seitz, K.W., Teske, A.P., and Baker, B.J., 2017. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome, 5(1):106. https://doi.org/
Dombrowski, N., Teske, A.P., and Baker, B.J., 2018. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nature Communications, 9(1):4999. https://doi.org/
Edgcomb, V.P., Beaudoin, D., Gast, R., Biddle, J.F., and Teske, A., 2011. Marine subsurface eukaryotes: the fungal majority. Environmental Microbiology, 13(1):172–183. https://doi.org/
Eme, L., Tamarit, D., Caceres, E.F., Stairs, C.W., De Anda, V., Schön, M.E., Seitz, K.W., Dombrowski, N., Lewis, W.H., Homa, F., Saw, J.H., Lombard, J., Nunoura, T., Li, W.-J., Hua, Z.-S., Chen, L.-X., Banfield, J.F., John, E.S., Reysenbach, A.-L., Stott, M.B., Schramm, A., Kjeldsen, K.U., Teske, A.P., Baker, B.J., and Ettema, T.J.G., 2023. Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes. Nature, 618(7967):992–999. https://doi.org/
Engelen, B., Nguyen, T., Heyerhoff, B., Kalenborn, S., Sydow, K., Tabai, H., Peterson, R.N., Wegener, G., and Teske, A., 2021. Microbial communities of hydrothermal Guaymas Basin surficial sediment profiled at 2 millimeter-scale resolution. Frontiers in Microbiology, 12.
https://doi.org/
Geller-McGrath, D., Konwar, K.M., Edgcomb, V.P., Pachiadaki, M., Roddy, J.W., Wheeler, T.J., and McDermott, J.E., 2024. Predicting metabolic modules in incomplete bacterial genomes with MetaPathPredict. eLife, 13:e85749. https://doi.org/
Heuer, V.B., Inagaki, F., Morono, Y., Kubo, Y., Maeda, L., Bowden, S., Cramm, M., Henkel, S., Hirose, T., Homola, K., Hoshino, T., Ijiri, A., Imachi, H., Kamiya, N., Kaneko, M., Lagostina, L., Manners, H., McClelland, H.-L., Metcalfe, K., Okutsu, N., Pan, D., Raudsepp, M.J., Sauvage, J., Schubotz, F., Spivack, A., Tonai, S., Treude, T., Tsang, M.-Y., Viehweger, B., Wang, D.T., Whitaker, E., Yamamoto, Y., and Yang, K., 2017. Site C0023. In Heuer, V.B., Inagaki, F., Morono, Y., Kubo, Y., Maeda, L., and the Expedition 370 Scientists, Temperature Limit of the Deep Biosphere off Muroto. Proceedings of the International Ocean Discovery Program, 370: College Station, TX (International Ocean Discovery Program). https://doi.org/
Heuer, V.B., Inagaki, F., Morono, Y., Kubo, Y., Spivack, A.J., Viehweger, B., Treude, T., Beulig, F., Schubotz, F., Tonai, S., Bowden, S.A., Cramm, M., Henkel, S., Hirose, T., Homola, K., Hoshino, T., Ijiri, A., Imachi, H., Kamiya, N., Kaneko, M., Lagostina, L., Manners, H., McClelland, H.-L., Metcalfe, K., Okutsu, N., Pan, D., Raudsepp, M.J., Sauvage, J., Tsang, M.-Y., Wang, D.T., Whitaker, E., Yamamoto, Y., Yang, K., Maeda, L., Adhikari, R.R., Glombitza, C., Hamada, Y., Kallmeyer, J., Wendt, J., Wörmer, L., Yamada, Y., Kinoshita, M., and Hinrichs, K.-U., 2020. Temperature limits to deep subseafloor life in the Nankai Trough subduction zone. Science, 370(6521):1230–1234. https://doi.org/
Hilz, H., Wiegers, U., and Adamietz, P., 1975. Stimulation of proteinase K action by denaturing agents: application to the isolation of nucleic acids and the degradation of ‘masked’ proteins. Eur J Biochem, 56(1):103-108. https://doi.org/
Hinkle, J.E., Chanton, J.P., Moynihan, M.A., Ruff, S.E., and Teske, A., preprint. Complex bacterial diversity of Guaymas Basin hydrothermal sediments revealed by synthetic long-read sequencing (LoopSeq). bioRxiv, Posted online 22 May 2024. https://doi.org/
Hinkle, J.E., Mara, P., Beaudoin, D.J., Edgcomb, V.P., and Teske, A.P., 2023. A PCR-based survey of methane-cycling Archaea in methane-soaked subsurface sediments of Guaymas Basin, Gulf of California. Microorganisms, 11(12):2956. https://doi.org/
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Lauer, A., Suzuki, M., Takai, K., Delwiche, M., Colwell, F.S., Nealson, K.H., Horikoshi, K., D’Hondt, S., and Jørgensen, B.B., 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean margin. Proceedings of the National Academy of Sciences of the United States of America, 103(8):2815–2820. https://doi.org/
Jiang, W., Liang, P., Wang, B., Fang, J., Lang, J., Tian, G., Jiang, J., and Zhu, T.F., 2015. Optimized DNA extraction and metagenomic sequencing of airborne microbial communities. Nature Protocols, 10(5):768–779. https://doi.org/
Kallmeyer, J., Smith, D.C., Spivack, A.J., and D’Hondt, S., 2008. New cell extraction procedure applied to deep subsurface sediments. Limnology and Oceanography: Methods, 6(6):236–245. https://doi.org/
Lagostina, L., Frandsen, S., MacGregor, B.J., Glombitza, C., Deng, L., Fiskal, A., Li, J., Doll, M., Geilert, S., Schmidt, M., Scholz, F., Bernasconi, S.M., Jørgensen, B.B., Hensen, C., Teske, A., and Lever, M.A., 2021. Interactions between temperature and energy supply drive microbial communities in hydrothermal sediment. Communications Biology, 4(1):1006. https://doi.org/
Lappé, M., and Kallmeyer, J., 2011. A cell extraction method for oily sediments. Frontiers in Microbiology, 2. https://doi.org/
Lever, M.A., and Teske, A.P., 2015. Diversity of methane-cycling archaea in hydrothermal sediment investigated by general and group-specific PCR primers. Applied and Environmental Microbiology, 81(4):1426–1441. https://doi.org/
Lizarralde, D., Teske, A., Höfig, T.W., González-Fernández, A., and IODP Expedition 385 Scientists, 2023. Carbon released by sill intrusion into young sediments measured through scientific drilling. Geology. https://doi.org/
Mara, P., Geller-McGrath, D., Edgcomb, V., Beaudoin, D., Morono, Y., and Teske, A., 2023a. Metagenomic profiles of archaea and bacteria within thermal and geochemical gradients of the Guaymas Basin deep subsurface. Nature Communications, 14(1):7768. https://doi.org/
Mara, P., Zhou, Y.-L., Teske, A., Morono, Y., Beaudoin, D., and Edgcomb, V., 2023b. Microbial gene expression in Guaymas Basin subsurface sediments responds to hydrothermal stress and energy limitation. The ISME Journal. https://doi.org/
McKay, L., Klokman, V.W., Mendlovitz, H.P., LaRowe, D.E., Hoer, D.R., Albert, D., Amend, J.P., and Teske, A., 2016. Thermal and geochemical influences on microbial biogeography in the hydrothermal sediments of Guaymas Basin, Gulf of California. Environmental Microbiology Reports, 8(1):150–161. https://doi.org/
Meyer, S., Wegener, G., Lloyd, K.G., Teske, A., Boetius, A., and Ramette, A., 2013. Microbial habitat connectivity across spatial scales and hydrothermal temperature gradients at Guaymas Basin. Frontiers in Microbiology, 4:207. https://doi.org/
Morono, Y., 2023. Accessing the energy-limited and sparsely populated deep biosphere: achievements and ongoing challenges of available technologies. Progress in Earth and Planetary Science, 10(1):18. https://doi.org/
Morono, Y., Terada, T., Hoshino, T., and Inagaki, F., 2014. Hot-alkaline DNA extraction method for deep-subseafloor archaeal communities. Applied and Environmental Microbiology, 80(6):1985–1994. https://doi.org/
Morono, Y., Terada, T., Kallmeyer, J., and Inagaki, F., 2013. An improved cell separation technique for marine subsurface sediments: applications for high-throughput analysis using flow cytometry and cell sorting. Environmental Microbiology, 15(10):2841–2879. https://doi.org/
Morono, Y., Teske, A., Galerne, C., Bojanova, D., Edgcomb, V., Meyer, N., Schubert, F., and Toffin, L. and the IODP Expedition 385 Scientists, 2022. Microbial cell distribution in the Guaymas Basin subseafloor biosphere, a young marginal rift basin with rich organics and steep temperature gradient. Presented at the EGU General Assembly, Vienna, Austria, 23–27 May 2022. https://doi.org/
Neumann, F., Negrete-Aranda, R., Harris, R.N., Contreras, J., Galerne, C.Y., Peña-Salinas, M.S., Spelz, R.M., Teske, A., Lizarralde, D., Höfig, T.W., and Scientists, E., 2023. Heat flow and thermal regime in the Guaymas Basin, Gulf of California: estimates of conductive and advective heat transport. Basin Research. https://doi.org/
Orsi, W., Biddle, J.F., and Edgcomb, V., 2013a. Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PloS One, 8(2):e56335. https://doi.org/
Orsi, W.D., Edgcomb, V.P., Christman, G.D., and Biddle, J.F., 2013b. Gene expression in the deep biosphere. Nature, 499(7457):205–208. https://doi.org/
Pachiadaki, M.G., Rédou, V., Beaudoin, D.J., Burgaud, G., and Edgcomb, V.P., 2016. Fungal and prokaryotic activities in the marine subsurface biosphere at Peru margin and Canterbury Basin inferred from RNA-based analyses and microscopy. Frontiers in Microbiology, 7. https://doi.org/
Parada, A.E., Needham, D.M., and Fuhrman, J.A., 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environmental Microbiology, 18(5):1403–1414. https://doi.org/
Parkes, R.J., Cragg, B., Roussel, E., Webster, G., Weightman, A., and Sass, H., 2014. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere-geosphere interactions. Marine Geology, 352:409–425. https://doi.org/
Parkes, R.J., Cragg, B.A., and Wellsbury, P., 2000. Recent studies on bacterial populations and processes in subseafloor sediments: a review. Hydrogeology Journal, 8(1):11–28. https://doi.org/
Quince, C., Lanzen, A., Davenport, R.J., and Turnbaugh, P.J., 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics, 12(1):38. https://doi.org/
Ramírez, G.A., Graham, D., and D’Hondt, S., 2018. Influence of commercial DNA extraction kit choice on prokaryotic community metrics in marine sediment. Limnology and Oceanography: Methods, 16(9):525–536. https://doi.org/
Ramírez, G.A., Paraskevi, V.M., Sehein, T., Wegener, G., Chambers, C.R., Joye, S.B., Peterson, R.N., Philippe, A., Burgaud, G., Edgcomb, V.P., and Teske, A.P. , 2021. Environmental factors shaping bacterial, archaeal and fungal community structure in hydrothermal sediments of Guaymas Basin, Gulf of California. PloS One. https://doi.org/
Salter, S.J., Cox, M.J., Turek, E.M., Calus, S.T., Cookson, W.O., Moffatt, M.F., Turner, P., Parkhill, J., Loman, N.J., and Walker, A.W., 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biology, 12:87. https://doi.org/
Seitz, K.W., Dombrowski, N., Eme, L., Spang, A., Lombard, J., Sieber, J.R., Teske, A.P., Ettema, T.J.G., and Baker, B.J., 2019. Asgard archaea capable of anaerobic hydrocarbon cycling. Nature Communications, 10(1):1822. https://doi.org/
Sørensen, K.B., and Teske, A., 2006. Stratified communities of active Archaea in deep marine subsurface sediments. Applied and Environmental Microbiology, 72(7):4596-4603. https://doi.org/
Su, L., Teske, A.P., MacGregor, B.J., McKay, L.J., Mendlovitz, H., Albert, D., Ma, Z., and Li, J., 2023. Thermal selection of microbial communities and preservation of microbial function in Guaymas Basin hydrothermal sediments. Applied and Environmental Microbiology, 89(3):e0001823. https://doi.org/
Suzuki, M.T., Taylor, L.T., and DeLong, E.F., 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Applied and Environmental Microbiology, 66(11):4605–4614. https://doi.org/
Tenzer, R., and Gladkikh, V., 2014. Assessment of density variations of marine sediments with ocean and sediment depths. The Scientific World Journal, 2014(1):823296. https://doi.org/
Teske, A., de Beer, D., McKay, L.J., Tivey, M.K., Biddle, J.F., Hoer, D., Lloyd, K.G., Lever, M.A., Røy, H., Albert, D.B., Mendlovitz, H.P., and MacGregor, B.J., 2016. The Guaymas Basin hiking guide to hydrothermal mounds, chimneys, and microbial mats: complex seafloor expressions of subsurface hydrothermal circulation. Frontiers in Microbiology, 7. https://doi.org/
Teske, A., Lizarralde, D., Höfig, T.W., Aiello, I.W., Ash, J.L., Bojanova, D.P., Buatier, M.D., Edgcomb, V.P., Galerne, C.Y., Gontharet, S., Heuer, V.B., Jiang, S., Kars, M.A.C., Khogenkumar Singh, S., Kim, J.-H., Koornneef, L.M.T., Marsaglia, K.M., Meyer, N.R., Morono, Y., Negrete-Aranda, R., Neumann, F., Pastor, L.C., Peña-Salinas, M.E., Pérez Cruz, L.L., Ran, L., Riboulleau, A., Sarao, J.A., Schubert, F., Stock, J.M., Toffin, L.M.A.A., Xie, W., Yamanaka, T., and Zhuang, G., 2021a. Expedition 385 summary. In Teske, A., Lizarralde, D., Höfig, T.W., and the Expedition 385 Scientists, Guaymas Basin Tectonics and Biosphere. Proceedings of the International Ocean Discovery Program, 385: College Station, TX (International Ocean Discovery Program). https://doi.org/
Teske, A., Lizarralde, D., Höfig, T.W., Aiello, I.W., Ash, J.L., Bojanova, D.P., Buatier, M.D., Edgcomb, V.P., Galerne, C.Y., Gontharet, S., Heuer, V.B., Jiang, S., Kars, M.A.C., Khogenkumar Singh, S., Kim, J.-H., Koornneef, L.M.T., Marsaglia, K.M., Meyer, N.R., Morono, Y., Negrete-Aranda, R., Neumann, F., Pastor, L.C., Peña-Salinas, M.E., Pérez Cruz, L.L., Ran, L., Riboulleau, A., Sarao, J.A., Schubert, F., Stock, J.M., Toffin, L.M.A.A., Xie, W., Yamanaka, T., and Zhuang, G., 2021b. Site U1545. In Teske, A., Lizarralde, D., Höfig, T.W., and the Expedition 385 Scientists, Guaymas Basin Tectonics and Biosphere. Proceedings of the International Ocean Discovery Program, 385: College Station, TX (International Ocean Discovery Program). https://doi.org/
Teske, A., Lizarralde, D., Höfig, T.W., Aiello, I.W., Ash, J.L., Bojanova, D.P., Buatier, M.D., Edgcomb, V.P., Galerne, C.Y., Gontharet, S., Heuer, V.B., Jiang, S., Kars, M.A.C., Khogenkumar Singh, S., Kim, J.-H., Koornneef, L.M.T., Marsaglia, K.M., Meyer, N.R., Morono, Y., Negrete-Aranda, R., Neumann, F., Pastor, L.C., Peña-Salinas, M.E., Pérez Cruz, L.L., Ran, L., Riboulleau, A., Sarao, J.A., Schubert, F., Stock, J.M., Toffin, L.M.A.A., Xie, W., Yamanaka, T., and Zhuang, G., 2021c. Site U1546. In Teske, A., Lizarralde, D., Höfig, T.W., and the Expedition 385 Scientists, Guaymas Basin Tectonics and Biosphere. Proceedings of the International Ocean Discovery Program, 385: College Station, TX (International Ocean Discovery Program). https://doi.org/
Teske, A., Lizarralde, D., Höfig, T.W., Aiello, I.W., Ash, J.L., Bojanova, D.P., Buatier, M.D., Edgcomb, V.P., Galerne, C.Y., Gontharet, S., Heuer, V.B., Jiang, S., Kars, M.A.C., Khogenkumar Singh, S., Kim, J.-H., Koornneef, L.M.T., Marsaglia, K.M., Meyer, N.R., Morono, Y., Negrete-Aranda, R., Neumann, F., Pastor, L.C., Peña-Salinas, M.E., Pérez Cruz, L.L., Ran, L., Riboulleau, A., Sarao, J.A., Schubert, F., Stock, J.M., Toffin, L.M.A.A., Xie, W., Yamanaka, T., and Zhuang, G., 2021d. Sites U1547 and U1548. In Teske, A., Lizarralde, D., Höfig, T.W., and the Expedition 385 Scientists, Guaymas Basin Tectonics and Biosphere. Proceedings of the International Ocean Discovery Program, 385: College Station, TX (International Ocean Discovery Program). https://doi.org/
Teske, A., Lizarralde, D., Höfig, T.W., Aiello, I.W., Ash, J.L., Bojanova, D.P., Buatier, M.D., Edgcomb, V.P., Galerne, C.Y., Gontharet, S., Heuer, V.B., Jiang, S., Kars, M.A.C., Khogenkumar Singh, S., Kim, J.-H., Koornneef, L.M.T., Marsaglia, K.M., Meyer, N.R., Morono, Y., Negrete-Aranda, R., Neumann, F., Pastor, L.C., Peña-Salinas, M.E., Pérez Cruz, L.L., Ran, L., Riboulleau, A., Sarao, J.A., Schubert, F., Stock, J.M., Toffin, L.M.A.A., Xie, W., Yamanaka, T., and Zhuang, G., 2021e. Site U1549. In Teske, A., Lizarralde, D., Höfig, T.W., and the Expedition 385 Scientists, Guaymas Basin Tectonics and Biosphere. Proceedings of the International Ocean Discovery Program, 385: College Station, TX (International Ocean Discovery Program). https://doi.org/
Teske, A., McKay, L.J., Ravelo, A.C., Aiello, I., Mortera, C., Núñez-Useche, F., Canet, C., et al., 2019. Characteristics and evolution of sill-driven off-axis hydrothermalism in Guaymas Basin – the Ringvent site. Scientific Reports, 9(1):13847. https://doi.org/
Teske, A., Wegener, G., Chanton, J.P., White, D., MacGregor, B., Hoer, D., de Beer, D., Zhuang, G., Saxton, M.A., Joye, S.B., Lizarralde, D., Soule, S.A., and Ruff, S.E., 2021f. Microbial communities under distinct thermal and geochemical regimes in axial and off-axis sediments of Guaymas Basin. Frontiers in Microbiology, 12:633649. https://doi.org/
Tighe, S., Afshinnekoo, E., Rock, T.M., McGrath, K., Alexander, N., McIntyre, A., Ahsanuddin, S., Bezdan, D., Green, S.J., Joye, S., Stewart Johnson, S., Baldwin, D.A., Bivens, N., Ajami, N., Carmical, J.R., Herriott, I.C., Colwell, R., Donia, M., Foox, J., Greenfield, N., Hunter, T., Hoffman, J., Hyman, J., Jorgensen, E., Krawczyk, D., Lee, J., Levy, S., Garcia-Reyero, N., Settles, M., Thomas, K., Gómez, F., Schriml, L., Kyrpides, N., Zaikova, E., Penterman, J., and Mason, C.E., 2017. Genomic methods and microbiological technologies for profiling novel and extreme environments for the Extreme Microbiome Project (XMP). Journal of Biomolecular Techniques, 28(1):31–39. https://doi.org/
Torti, A., Lever, M.A., and Jørgensen, B.B., 2015. Origin, dynamics, and implications of extracellular DNA pools in marine sediments. Marine Genomics, 24:185–196. https://doi.org/
Zhou, J., Bruns, M.A., and Tiedje, J.M., 1996. DNA recovery from soils of diverse composition. Applied and Environmental Microbiology, 62(2):316-322. https://doi.org/